- Current
- Browse
- Collections
-
For contributors
- For Authors
- Instructions to authors
- Article processing charge
- e-submission
- For Reviewers
- Instructions for reviewers
- How to become a reviewer
- Best reviewers
- For Readers
- Readership
- Subscription
- Permission guidelines
- About
- Editorial policy
Articles
- Page Path
- HOME > Diabetes Metab J > Volume 44(6); 2020 > Article
-
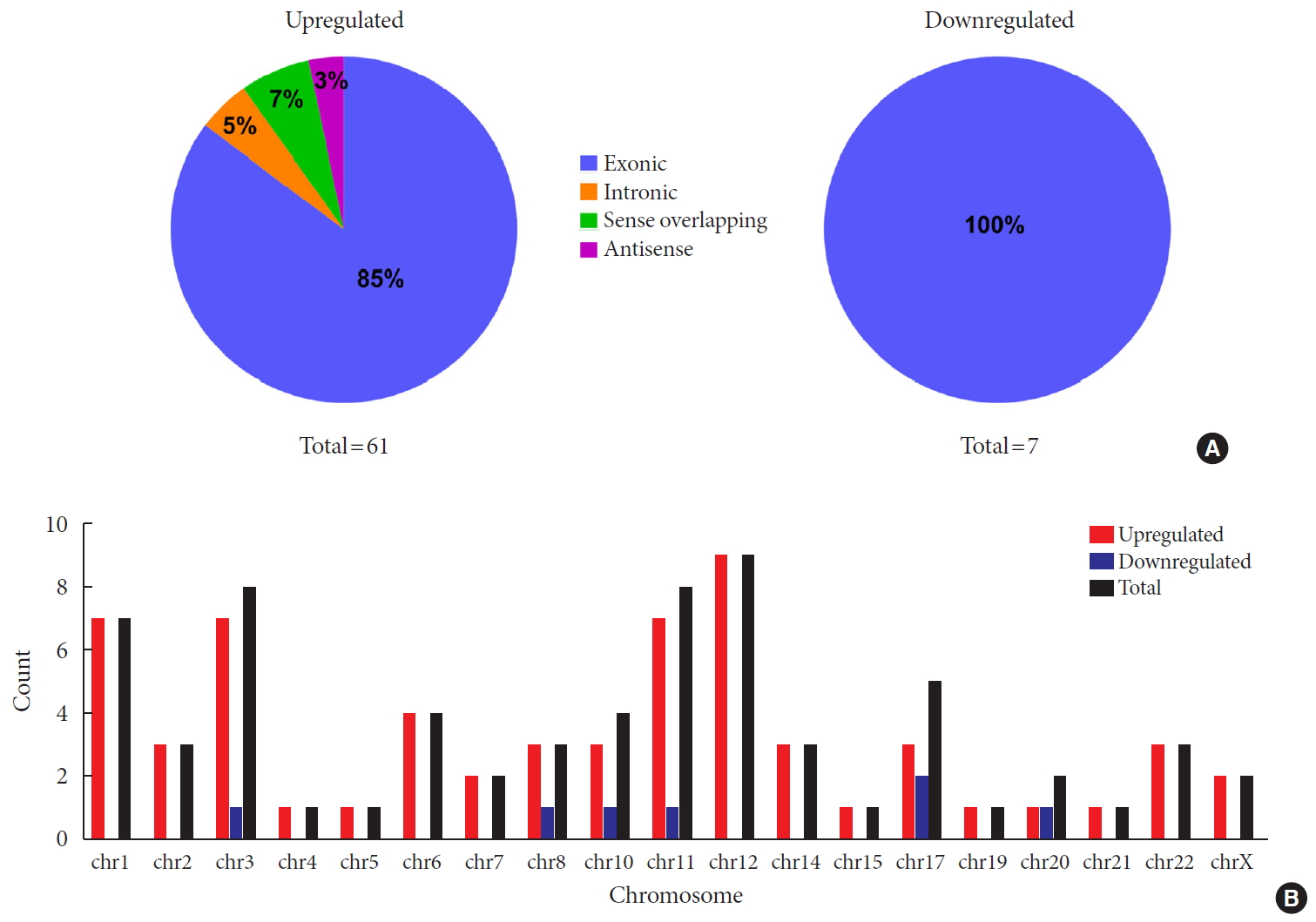
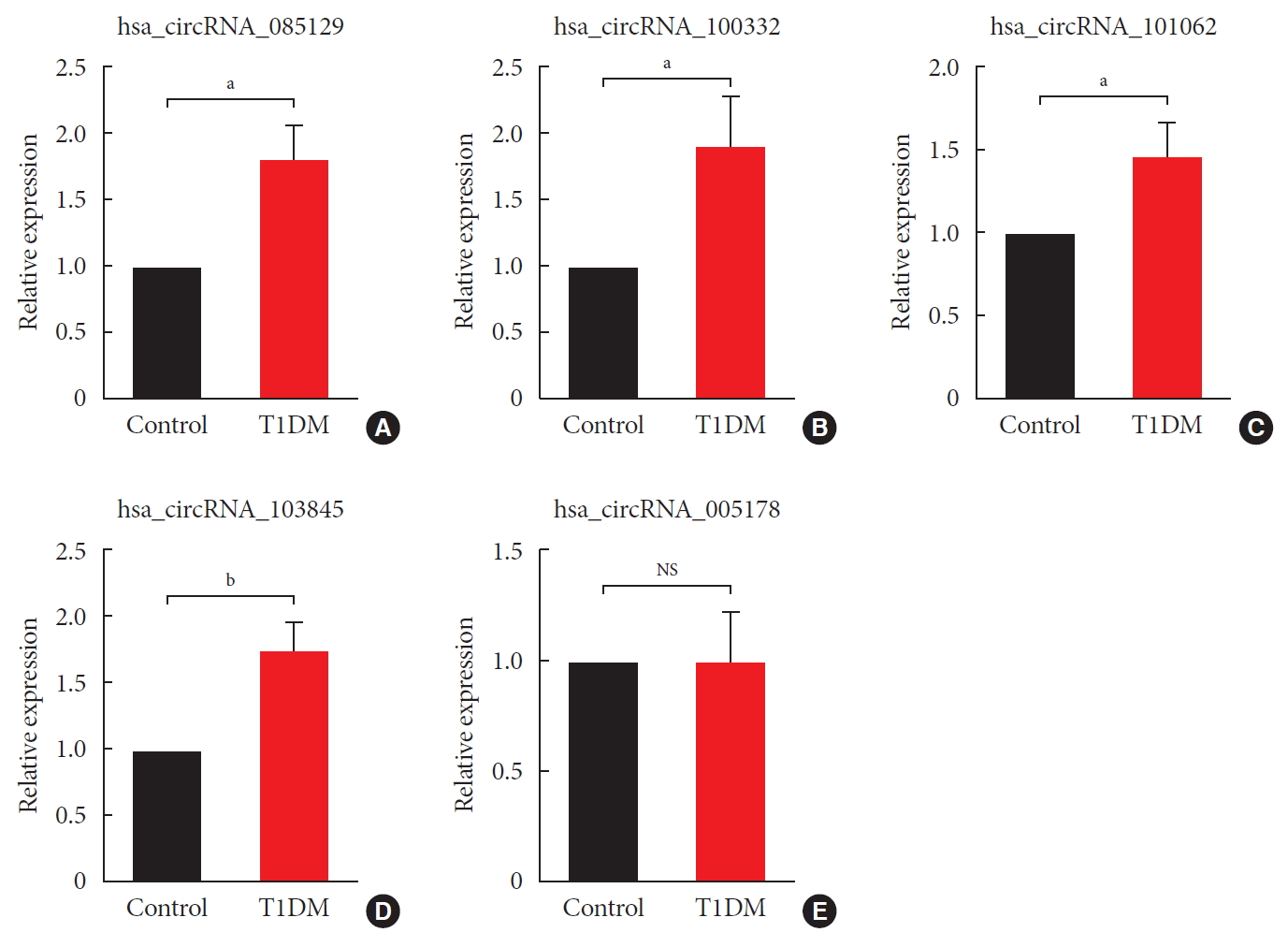
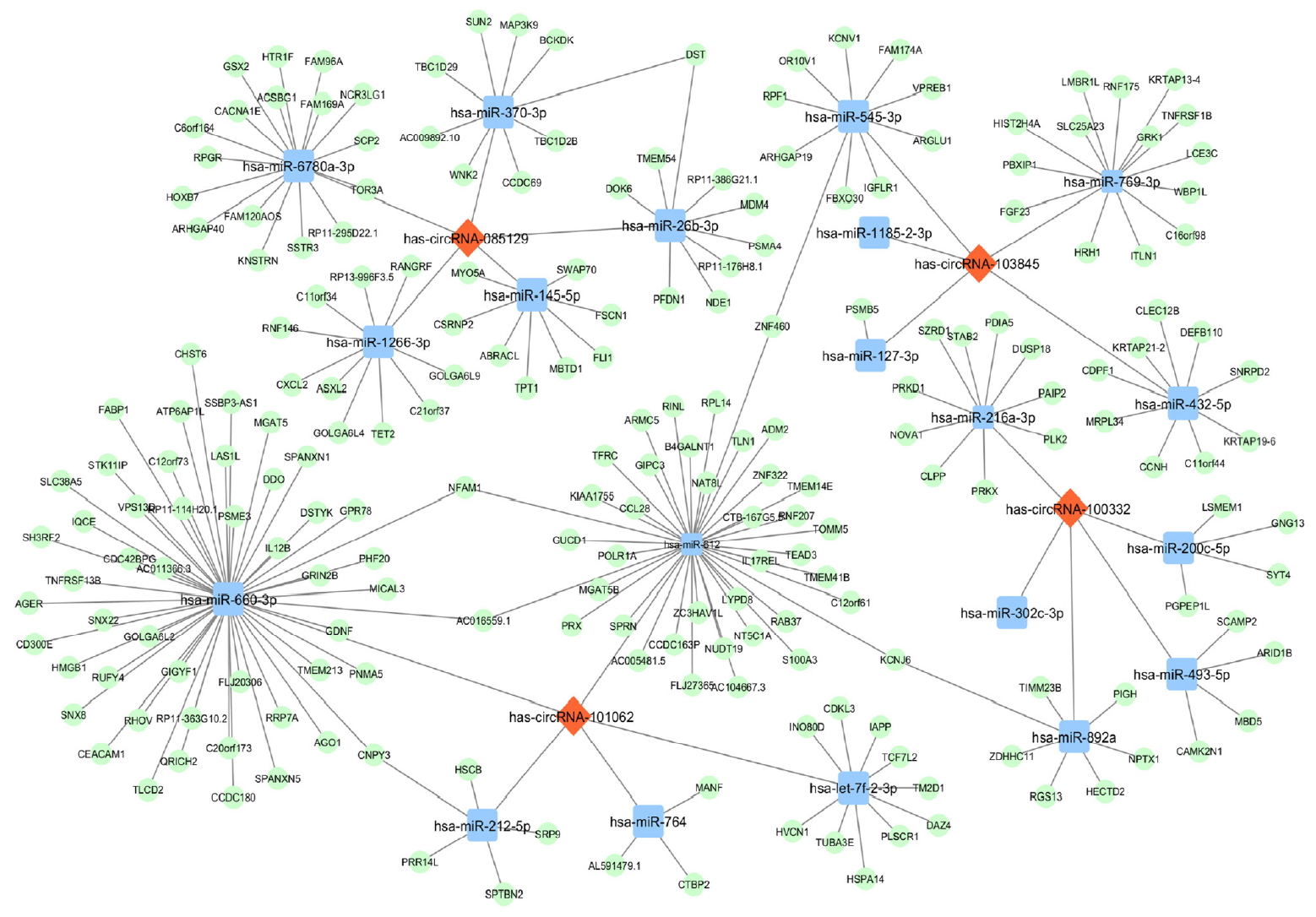
Original ArticleType 1 Diabetes Differential Profile of Plasma Circular RNAs in Type 1 Diabetes Mellitus
-
Yangyang Li1
 , Ying Zhou2, Minghui Zhao1, Jing Zou2, Yuxiao Zhu2, Xuewen Yuan3, Qianqi Liu3, Hanqing Cai1, Cong-Qiu Chu4,5
, Ying Zhou2, Minghui Zhao1, Jing Zou2, Yuxiao Zhu2, Xuewen Yuan3, Qianqi Liu3, Hanqing Cai1, Cong-Qiu Chu4,5 , Yu Liu2
, Yu Liu2
-
Diabetes & Metabolism Journal 2020;44(6):854-865.
DOI: https://doi.org/10.4093/dmj.2019.0151
Published online: July 13, 2020
1Department of Endocrinology, The Second Hospital of Jilin University, Changchun, China
2Department of Endocrinology, Sir Run Run Hospital, Nanjing Medical University, Nanjing, China
3Department of Endocrinology, Children's Hospital of Nanjing Medical University, Nanjing, China
4Division of Arthritis and Rheumatic Diseases, Oregon Health & Science University School of Medicine, Portland, OR, USA
5Section of Rheumatology, VA Portland Health Care System, Portland, OR, USA
- Corresponding author: Yu Liu. Department of Endocrinology, Sir Run Run Hospital, Nanjing Medical University, Nanjing 211166, China. drliuyu@njmu.edu.cn
- Corresponding author: Cong-Qiu Chu. Division of Arthritis and Rheumatic Diseases, Oregon Health & Science University School of Medicine, Portland, OR 97239, USA. chuc@ohsu.edu
Copyright © 2020 Korean Diabetes Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Non-coding RNAs and exosomal non-coding RNAs in diabetic retinopathy: A narrative review
Yuhong Zhong, Juan Xia, Li Liao, Mohammad Reza Momeni
International Journal of Biological Macromolecules.2024; 259: 128182. CrossRef - Circular RNAs: Potential biomarkers and therapeutic targets for autoimmune diseases
Ren-Jie Zhao, Wan-Ying Zhang, Xing-Xing Fan
Heliyon.2024; 10(1): e23694. CrossRef - Research progress of circular RNA molecules in aging and age-related diseases
Zhidan Zhang, Yuling Huang, AYao Guo, Lina Yang
Ageing Research Reviews.2023; 87: 101913. CrossRef - CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders
Yuka Ikeda, Sae Morikawa, Moeka Nakashima, Sayuri Yoshikawa, Kurumi Taniguchi, Haruka Sawamura, Naoko Suga, Ai Tsuji, Satoru Matsuda
Non-Coding RNA.2023; 9(2): 23. CrossRef - Decrypting the circular RNAs does a favor for us: Understanding, diagnosing and treating diabetes mellitus and its complications
Zi Li, Yuanyuan Ren, Ziwei Lv, Man Li, Yujia Li, Xiaobin Fan, Yuyan Xiong, Lu Qian
Biomedicine & Pharmacotherapy.2023; 168: 115744. CrossRef - Circular RNA PIP5K1A Promotes Glucose and Lipid Metabolism Disorders and Inflammation in Type 2 Diabetes Mellitus
Ge Song, YiQian Zhang, YiHua Jiang, Huan Zhang, Wen Gu, Xiu Xu, Jing Yao, ZhengFang Chen
Molecular Biotechnology.2023;[Epub] CrossRef - Hsa_circRNA_405498 and hsa_circRNA_100033 Serve as Potential Biomarkers for Differential Diagnosis of Type 1 Diabetes
Ziwei Zhang, Shuoming Luo, Zilin Xiao, Wenfeng Yin, Xiajie Shi, Hongzhi Chen, Zhiguo Xie, Zhenqi Liu, Xia Li, Zhiguang Zhou
The Journal of Clinical Endocrinology & Metabolism.2023;[Epub] CrossRef - Circular RNA PIP5K1A act as microRNA-552-3p sponge to regulates inflammation, oxidative damage in glucolipotoxicity-induced pancreatic INS-1 β-cells via Janus kinase 1
Lei Ren
Bioengineered.2022; 13(3): 5724. CrossRef - Circular RNAs in diabetes mellitus and its complications
Wenqi Fan, Haipeng Pang, Zhiguo Xie, Gan Huang, Zhiguang Zhou
Frontiers in Endocrinology.2022;[Epub] CrossRef - Type 1 Diabetes Mellitus-Related circRNAs Regulate CD4+ T Cell Functions
Jianni Chen, Guanfei Jia, Xue Lv, Shufa Li, Christos K. Kontos
BioMed Research International.2022; 2022: 1. CrossRef - An intriguing role of circular RNA in insulin resistance and endothelial dysfunction: the future perspectives
Monisha Prasad, Selvaraj Jayaraman, Vishnu Priya Veeraraghavan
Hypertension Research.2022; 45(11): 1843. CrossRef - Circular RNAs in Diabetic Nephropathy: Updates and Perspectives
Miao Liu, Junli Zhao
Aging and disease.2022; 13(5): 1365. CrossRef - CircRNAs: Key molecules in the prevention and treatment of ischemic stroke
Zeyu Liu, Yanhong Zhou, Jian Xia
Biomedicine & Pharmacotherapy.2022; 156: 113845. CrossRef - Pro-Inflammatory Cytokines Promote the Transcription of Circular RNAs in Human Pancreatic β Cells
Simranjeet Kaur, Caroline Frørup, Aashiq H. Mirza, Tina Fløyel, Reza Yarani, Maikel L. Colli, Jesper Johannesen, Joachim Størling, Decio L. Eizirik, Flemming Pociot
Non-Coding RNA.2022; 8(5): 69. CrossRef - Differential Expression and Bioinformatics Analysis of Plasma-Derived Exosomal circRNA in Type 1 Diabetes Mellitus
Haipeng Pang, Wenqi Fan, Xiajie Shi, Shuoming Luo, Yimeng Wang, Jian Lin, Yang Xiao, Xia Li, Gan Huang, Zhiguo Xie, Zhiguang Zhou, Jinhui Liu
Journal of Immunology Research.2022; 2022: 1. CrossRef - Circular RNAs in diabetes and its complications: Current knowledge and future prospects
Wenfeng Yin, Ziwei Zhang, Zilin Xiao, Xia Li, Shuoming Luo, Zhiguang Zhou
Frontiers in Genetics.2022;[Epub] CrossRef - Circular RNA in autoimmune diseases: special emphasis on regulation mechanism in RA and SLE
Yurong Huang, Qiuyun Xue, Chenglong Cheng, Yuting Wang, Xiao Wang, Jun Chang, Chenggui Miao
Journal of Pharmacy and Pharmacology.2022;[Epub] CrossRef - Emerging roles of circular RNAs in systemic lupus erythematosus
Xin Wang, Rui Ma, Weimin Shi, Zhouwei Wu, Yuling Shi
Molecular Therapy - Nucleic Acids.2021; 24: 212. CrossRef - Understanding Competitive Endogenous RNA Network Mechanism in Type 1 Diabetes Mellitus Using Computational and Bioinformatics Approaches
Xuanzi Yi, Xu Cheng
Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy.2021; Volume 14: 3865. CrossRef
- Figure
- Related articles
-
- Navigating the Seas of Glycemic Control: The Role of Continuous Glucose Monitoring in Type 1 Diabetes Mellitus
- Change Profiles and Functional Targets of MicroRNAs in Type 2 Diabetes Mellitus Patients with Obesity
- Blood Pressure Target in Type 2 Diabetes Mellitus
- Implication of Sex Differences in Visceral Fat for the Assessment of Incidence Risk of Type 2 Diabetes Mellitus
- Sex Differences of Visceral Fat Area and Visceral-to-Subcutaneous Fat Ratio for the Risk of Incident Type 2 Diabetes Mellitus

 KDA
KDA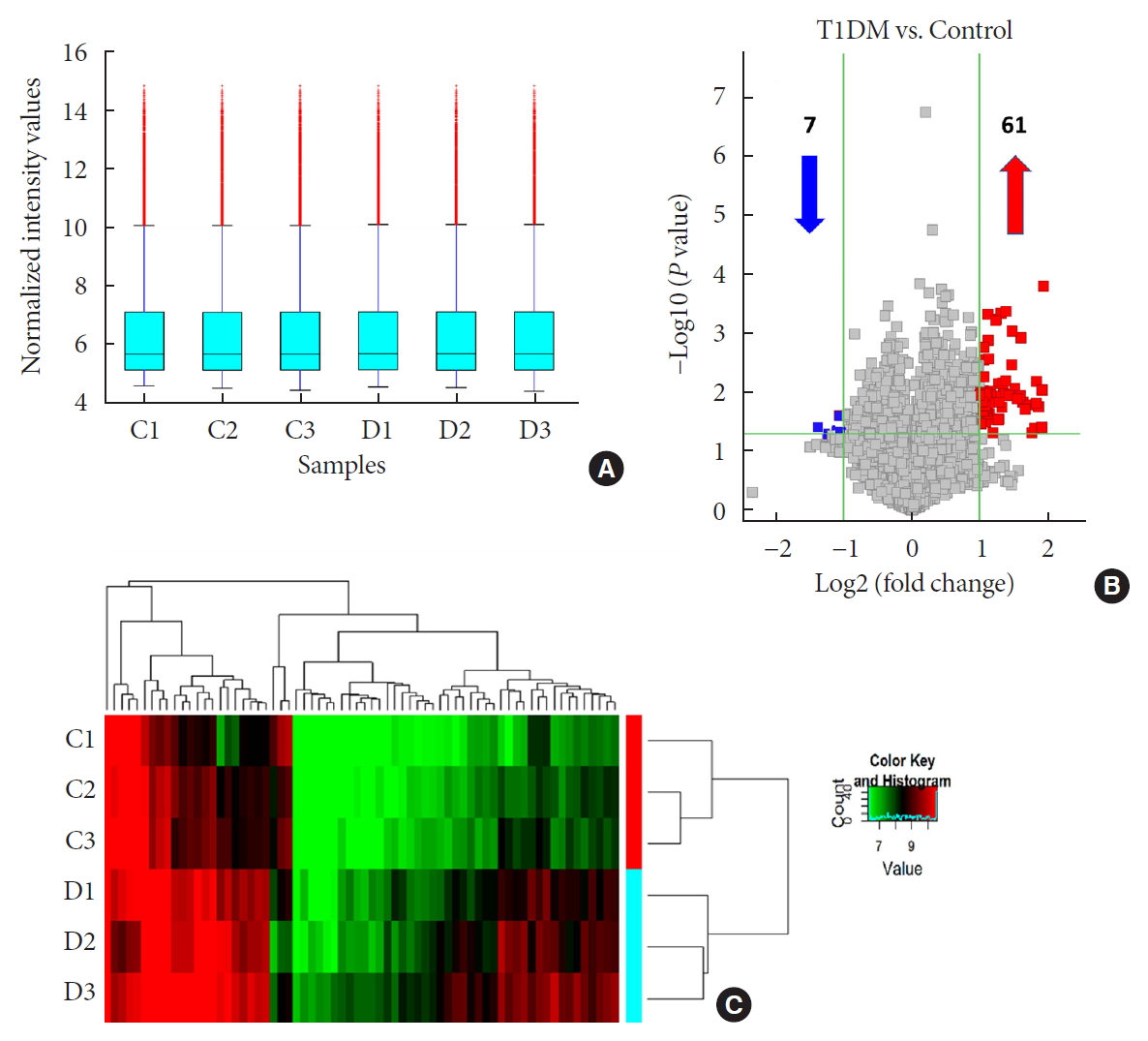

 PubReader
PubReader ePub Link
ePub Link Cite
Cite