- Current
- Browse
- Collections
-
For contributors
- For Authors
- Instructions to authors
- Article processing charge
- e-submission
- For Reviewers
- Instructions for reviewers
- How to become a reviewer
- Best reviewers
- For Readers
- Readership
- Subscription
- Permission guidelines
- About
- Editorial policy
Articles
- Page Path
- HOME > Diabetes Metab J > Volume 35(4); 2011 > Article
-
Original ArticleEffects of Sulfonylureas on Peroxisome Proliferator-Activated Receptor γ Activity and on Glucose Uptake by Thiazolidinediones
- Kyeong Won Lee, Yun Hyi Ku, Min Kim, Byung Yong Ahn, Sung Soo Chung, Kyong Soo Park
-
Diabetes & Metabolism Journal 2011;35(4):340-347.
DOI: https://doi.org/10.4093/dmj.2011.35.4.340
Published online: August 31, 2011
Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea.
- Corresponding author: Kyong Soo Park. Department of Internal Medicine & WCU Department of Molecular Medicine and Biopharmaceutical Sciences, Seoul National University College of Medicine, 28 Yeongeon-dong, Jongno-gu, Seoul 110-744, Korea. kspark@snu.ac.kr
• Received: November 10, 2010 • Accepted: May 10, 2011
Copyright © 2011 Korean Diabetes Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
-
Background
- Sulfonylurea primarily stimulates insulin secretion by binding to its receptor on the pancreatic β-cells. Recent studies have suggested that sulfonylureas induce insulin sensitivity through peroxisome proliferator-activated receptor γ (PPARγ), one of the nuclear receptors. In this study, we investigated the effects of sulfonylurea on PPARγ transcriptional activity and on the glucose uptake via PPARγ.
-
Methods
- Transcription reporter assays using Cos7 cells were performed to determine if specific sulfonylureas stimulate PPARγ transactivation. Glimepiride, gliquidone, and glipizide (1 to 500 µM) were used as treatment, and rosiglitazone at 1 and 10 µM was used as a control. The effects of sulfonylurea and rosiglitazone treatments on the transcriptional activity of endogenous PPARγ were observed. In addition, 3T3-L1 adipocytes were treated with rosiglitazone (10 µM), glimepiride (100 µM) or both to verify the effect of glimepiride on rosiglitazone-induced glucose uptake.
-
Results
- Sulfonylureas, including glimepiride, gliquidone and glipizide, increased PPARγ transcriptional activity, gliquidone being the most potent PPARγ agonist. However, no additive effects were observed in the presence of rosiglitazone. When rosiglitazone was co-treated with glimepiride, PPARγ transcriptional activity and glucose uptake were reduced compared to those after treatment with rosiglitazone alone. This competitive effect of glimepiride was observed only at high concentrations that are not achieved with clinical doses.
-
Conclusion
- Sulfonylureas like glimepiride, gliquidone and glipizide increased the transcriptional activity of PPARγ. Also, glimepiride was able to reduce the effect of rosiglitazone on PPARγ agonistic activity and glucose uptake. However, the competitive effect does not seem to occur at clinically feasible concentrations.
- Insulin resistance is a characteristic feature in the pathogenesis of type 2 diabetes mellitus [1-3]. Thiazolidinediones (TZDs) are a class of antidiabetic agents that improve peripheral insulin resistance and lead to lower fasting and postprandial glucose levels, as well as circulating insulin levels [3]. TZDs bind to peroxisome proliferator-activated receptor γ (PPARγ) within the cell nucleus and activate the transcription of several specific genes which result in glycemic control, increase in high density lipoprotein, reduction of triglyceride, free fatty acids and small, dense low density lipoprotein, inhibition of tumoral angiogenesis, anti-inflammatory effects and adipose cell differentiation [4,5]. PPARγ is a member of the nuclear receptor superfamily and functions as a heterodimer with retinoid X receptors (RXRs) [4,6]. The C-terminal region of PPARγ, called ligand binding domain, is dimerized with RXR and contains the major transcriptional activation domain, termed the activation-function 2 (AF2) domain. This region is important for the docking of coactivators and forms a ligand-binding pocket through conformational change [4]. Also, the N-terminal of PPARγ has transcriptional activity and leads to diverse biological actions [4,7].
- Various substances have been suggested to be natural ligands for PPARγ, such as fatty acids and eicosanoids [8], components of oxidized low-density lipoproteins [9] and nitrolinoleic acid [10,11]. Additionally, some synthetic compounds, including TZDs, some non-steroidal anti-inflammatory drugs [12] and telmisartan, an angiotensin II receptor antagonist [13,14], showed a partial agonistic effect to PPARγ. Recently, investigators demonstrated that some antidiabetic sulfonylureas, whose main mechanism in glycemic control is to stimulate insulin secretion by binding to the sulfonylurea receptor of pancreatic β-cells, played roles as PPAR agonists [15-18].
- In the present work, we compared PPARγ activating properties of sulfonylureas to those of rosiglitazone and also investigated their effects on PPARγ activity when combined with rosiglitazone.
INTRODUCTION
- Materials
- Rosiglitazone was purchased from Cayman Chemical (Ann Arbor, MI, USA), gliquidone from Apin Chemicals (Abingdon, UK), and glipizide from Sigma-Aldrich (Louis, MO, USA). Glimepiride was a kind gift from the Handok Co. (Seoul, Korea). Agents were dissolved in dimethyl sulfoxide (Sigma, St. Louis, MO, USA).
- Cell culture
- COS7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). 3T3-L1 preadipocytes were cultured in DMEM with 10% calf serum. Two days after 3T3-L1 cells had reached confluence, differentiation was induced by treating the cells with 10% FBS-supplemented DMEM containing 0.5 nM 3-isobutyl-1-methylxanthine (IBMX), 0.25 µM dexamethasone and 5 µg/mL insulin for 48 hours. Cells were refed in DMEM with 10% FBS and 1 µg/mL insulin for the following two days and were then maintained in DMEM with 10% FBS for the following four days.
- Construction of plasmids
- The Gal4 DNA binding domain fused expression vectors encoding the deletion mutant of mouse PPARγ2 were prepared by subcloning the corresponding cDNAs into a pM vector. The cDNAs encoding the full-length DNA (WT, amino acid 1-505), the activation function-1 domain (AF1, amino acid 1-138), the ligand binding domain-deleted construct (ΔLBD, amino acid 1-311) and only the ligand binding domain (LBD, amino acid 203-505) of the mouse PPARγ type 2 were generated using polymerase chain reaction with oligonucleotide primers. Primer sets were as follows: WT, AF1, and ΔLBD sense primer, 5'-agt cga ctg ggt gaa act ctg gga gat tc-3'; LBD sense primer, 5'-atg tcg acg gat gtc tca caa tgc cat cag g-3'; WT and LBD antisense primer, 5'-ggt cta gac ggg tgg gac ttt cct gc-3'; AF1 antisense primer, 5'-cct cta gac tca atg gcc atg agg-3'; ΔLBD antisense primer, 5'-att cta gat tga aaa att cgg atg gcc ac-3'. The amplicons were ligated into the pM vector containing the Gal4 DNA binding domain at the N-terminal using the SalI and XbaI sites. Gal4-responsive tk-Luc reporters (Gal4 tk-Luc) were used to evaluate the transcriptional activity of PPARγ, and β-galactosidase (pCMV-β-gal) was used to normalize the transient transfection efficiency.
- Transient transfection, treatment and reporter assay
- COS7 cells in 12-well plates were transfected with Gal4 tk-Luc (0.1 µg), pCMV-β-gal (0.1 µg) and pM-PPARγ constructs (0.03 µg) using LipofectAMIN Plus (Invitrogen, Carlsbad, CA, USA). Cells were incubated in DMEM supplemented with 10% FBS and were treated with glimepiride, gliquidone, glipizide and rosiglitazone at the indicated doses for 24 hours. In order to identify the concentration-dependent activation of PPARγ by sulfonylureas, 1, 10, and 100 µM of glimepiride and 1, 10, and 30 µM of gliquidone were used. Luciferase activity was determined using the Luciferase Assay System Kit (Promega, Madison, WI, USA) and Lumat LB9507 (Berthold, Bad Wildbad, Germany). Luciferase activity was normalized according to β-galactosidase activity.
- Glucose uptake
- 3T3-L1 adipocytes were treated with rosiglitazone (10 µM), glimepiride (100 µM) or both for 48 hours. After the addition of insulin (100 nM) to the medium for 30 minutes, cells were washed with salt-HEPES buffer (4.7 mM KCl, 130 mM NaCl, 1.25 mM CaCl2, 2.5 mM NaH2PO4, 1.2 mM MgSO4, and 10 mM HEPES). Cells were incubated in salt-HEPES buffer containing 0.2 µCi of [3H]deoxyglucose for 15 minutes. Uptake was terminated via five rapid washes with cold phosphate-buffered saline (PBS). Cells were lysed by 0.5N NaOH and neutralized by HCl. The radioactivity was determined using liquid scintillation counting in a β-counter and normalized according to total protein level.
- Statistics
- SPSS version 12.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. The data are expressed as mean±standard error. The differences between the means were calculated using the Mann-Whitney U-test. A P value less than 0.05 denoted the presence of a statistically significant difference.
METHODS
- PPARγ transcriptional activity by sulfonylureas
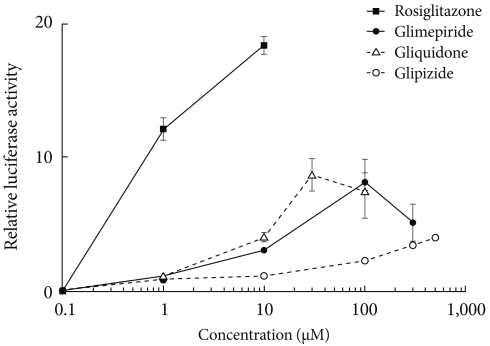
- Transcription reporter assays were used to determine the effects of sulfonylureas including glimepiride, gliquidone, and glipizide on the transcriptional activity of PPARγ. Each sulfonylurea was tested at the following concentrations: glimepiride, 1, 10, 100, and 300 µM; gliquidone, 1, 10, 30, and 100 µM; glipizide, 1, 10, 100, 300, and 500 µM and rosiglitazone, 1 and 10 µM. We obtained the peak concentration at which glimepiride and gliquidone induced the highest transcriptional activity; gliquidone, 30 µM and glimepiride, 100 µM. Glipizide reached saturation at concentrations greater than 500 µM, therefore, tests were performed at lower concentrations, although we did not obtain the peak value. All agents except glipizide significantly increased PPARγ agonistic activity at the indicated concentrations. Glimepiride and gliquidone increased PPARγ transcriptional activity at 1 µM, by 3-4 times at 10 µM, and by nearly ten times at higher concentrations. All of these increases were statistically significant, although they were lower than that of rosiglitazone. In the case of glipizide, no agonistic effect at 1 µM was observed, but there was a mild effect at concentrations greater than 10 µM (Fig. 1).
- Target region of sulfonylureas in PPARγ
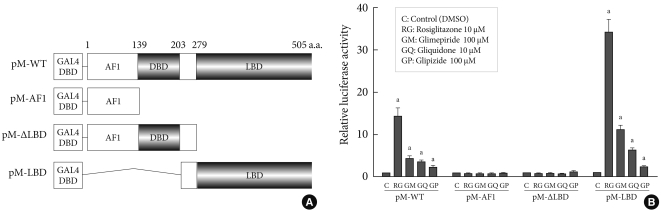
- In order to identify the target domain of sulfonylureas in PPARγ, we prepared three constructs from wild type PPARγ (pM-WT). One was the AF1 region (pM-AF1), another lacked the ligand binding domain (pM-ΔLBD), and the other was the ligand binding domain (pM-LBD) (Fig. 2A). The increment of PPARγ transcriptional activity caused by sulfonylureas disappeared when pM-AF1 or pM-ΔLBD was transfected, regardless of the type of sulfonylurea, whereas it was maintained in presence of pM-LBD (Fig. 2B). Based on these observations, it was suggested that the effect of sulfonylureas, like thiazolidinediones, on the PPARγ transcriptional activity involves binding to the ligand binding domain of PPARγ.
- Combinational treatment of sulfonylureas and rosiglitazone
- We examined the effects of rosiglitazone in addition to each sulfonylurea on the PPARγ transcriptional activity. When glimepiride (100 µM), gliquidone (10 µM), or glipizide (100 µM) was administered with rosiglitazone (1 µM), sulfonylureas did not provide any additive effects on the PPARγ transcriptional activity of the cells compared to that achieved with rosiglitazone alone. Interestingly, glimepiride reduced the effect of rosiglitazone on PPARγ transcriptional activity by about 50 percent, which was statistically significant, while gliquidone and glipizide showed no depletive influence (Fig. 3A).
- Because glimepiride and gliquidone were more effective than was glipizide for stimulation of PPARγ transcriptional activity, we investigated the effects of combination treatments with rosiglitazone in the presence of different sulfonylurea concentrations using glimepiride and gliquidone. In the combined treatment, glimepiride inhibited the effect of rosiglitazone in a dose-dependent manner. Under constant rosiglitazone treatment at a dose of 1 µM, when treated with glimepiride 1 µM, luciferase activity showed no change compared to that of the control; at 10 µM, luciferase activity seemed to be lowered, but the effect was not statistically significant. In contrast, when combined with 100 µM glimepiride, the reduction in activity was significantly greater; gliquidone did not show any difference in luciferase activity at any concentration, although gliquidone was similar or more effective than glimepiride at the indicated dose (Fig. 3B).
- Effect of glimepiride on glucose uptake by thiazolidinediones
- We evaluated the effects of rosiglitazone or glimepiride monotherapy and the combined therapy of the two on insulin-stimulated glucose uptake in adipocytes. Each monotherapy and the combination therapy produced an increase in glucose uptake. However, the glucose uptake in co-treatment was less than that of rosiglitazone monotherapy (Fig. 4). It is supposed that the effect of rosiglitazone on glucose uptake was inhibited by glimepiride, and this seems to be related to the effect of PPARγ transcriptional activity.
RESULTS
- There has been increasing interest in the effects of sulfonylurea on PPARγ transcriptional activity. The major findings of our experiments showed that sulfonylureas increased PPARγ transcriptional activity, and glimepiride showed dose-dependent competition with rosiglitazone for PPARγ, whereas gliquidone and glipizide did not exhibit competitive behaviors.
- It has been reported that some sulfonylureas enhance PPARγ transcriptional activity and, thus, target both sulfonylurea receptors in the membranes of pancreatic β-cells and PPARγ in the nuclei of adipocytes [15-18]. Arrault et al. [17] explored the PPARγ-activating properties of a series of eight sulfonylureas using transfection experiments with 293T cells and rosiglitazone as a reference PPARγ agonist. They found that second generation sulfonylureas, including glimepiride, gliquidone, and glibenclamide, stimulated PPARγ-activating properties, but first generation agents, such as tolbutamide, chlorpropamide, tolazamide and gliclazide, in addition to the second generation agent glipizide, did not [17]. Their findings were consistent with our results and with previously published data by Fukuen et al. [15], with the exception of the glipizide results. In the present study and the work of Fukuen et al. [15], glipizide increased the PPARγ transcriptional activity.
- Glipizide is known to exhibit a slightly different binding mode compared to those of glimepiride, gliquidone and glibenclamide [17,18]; glipizide binds PPARγ more weakly than do the others. The strength of this binding is determined by the hydrogen bonds (H-bond) formed between the docked ligand and the amino acids in PPARγ which form the binding pocket within the active site. The H-bonding pattern of glipizide is different from those of these other compounds [17]. Compared to glimepiride and gliquidone, which form more than two hydrogen bonds with the PPARγ ligand binding pocket, glipizide forms only one hydrogen bond and, thus, functions as a weak binder [17]. In this respect, glipizide showed weaker PPARγ agonistic activity than did the other second generation sulfonylureas. Additionally, glipizide may activate PPARγ in a tissue-specific manner; it showed PPARγ agonistic activity in CV-1 cells [18] and Cos7 cells but not in 293T cells [17].
- Other authors have observed PPARγ agonistic activity of glimepiride at 1 and 10 µM [15,16] similar to our results. The mean maximal plasma concentration (Cmax) values for glimepiride can reach 1 µM when patients are treated with the suggested maximum daily does of 8 mg [19,20]. For gliquidone, when treated with a 30 mg dose, Cmax is 1.2 µM, with a range from 0.2 to 0.4 µM [21]. In our experiment, gliquidone showed a PPARγ agonistic effect even when treated with 1 µM. Based on this result, glimepiride and gliquidone seem to be able to activate PPARγ at pharmacological concentrations. In addition, glimepiride, a third generation sulfonylurea, demonstrated the ability to improve insulin sensitivity in a human study [22]. Insulin resistance estimated using homeostatic model assessment-insulin resistance (HOMA-IR) was significantly reduced in subjects with type 2 diabetes who took glimepiride, although glycemic control measured using HbA1c was unchanged [22]. Also, gliquidone was shown to be as potent as pioglitazone for inducing PPARγ target gene expression [18].
- In the case of glipizide, which was able to weakly activate PPARγ at a dose of 10 µM in our experiment, Cmax values are 1.0±0.3 µM in subjects treated with a 5 mg dose [23]. Even in subjects administered the maximum daily dose of 40 mg, the concentration able to induce PPARγ agonistic activity is not achieved.
- One of the novel findings of this study was that glimepiride reduced the effect of rosiglitazone at high concentrations, but gliquidone or glipizide did not. In our results, glimepiride significantly inhibits the effect of rosiglitazone on PPARγ transcriptional activity in a dose-dependent manner, and cell viability was not affected in the tested dose range (data not shown). Our results are consistent with the results of a previous report [15].
- Fukuen and co-workers performed competitive binding assays using full-length PPARγ2 and [3H] rosiglitazone. The concentration-dependent displacement of [3H] rosiglitazone by glimepiride was observed; therefore, they concluded that glimepiride is in competition with rosiglitazone and activates PPARγ through direct association and could be considered as a partial agonist for PPARγ [15]. In the case of gliquidone, although it had stronger agonistic effects than did glimepiride, it did not reduce the effect of rosiglitazone even at 30 µM, the highest concentration tested.
- The explanation for these differences is not clear. It is known that the TZD head-group forms hydrogen bonds with the PPARγ residues which form the loop structure within the active site [17,24]. Glimepiride generated H-bonds with Ser289, Ser342, and the Gly284, whereas gliquidone interacted with Gln286 and Ser342 [17]. Rosiglitazone and glimepiride shared Ser289, but gliquidone did not. Glipizide, which showed no competitive effect against rosiglitazone, was a weak binder compared to the other sulfonylureas. Further investigation about the exact mechanism is needed.
- Though glimepiride inhibits the effect of rosiglitazone at a dose of 100 µM, competition between glimepiride and rosiglitazone is not expected in clinical practice since Cmax can be 1 µM at the maximum daily dose of glimepiride (8 mg).
- Although sulfonylurea drugs produce a glucose-lowering effect by stimulating insulin secretion in the pancreatic β-cells in an ATP-sensitive, K+ channel-dependent manner; they have also been proposed to have peripheral effects as an insulin sensitizer [22,25-27]. Glimepiride, the most recently developed sulfonylurea agent, induces glucose uptake in extrapancreatic tissue not only by increasing insulin secretion in the pancreatic β-cells, but also by stimulating GLUT1 and GLUT4 translocations in both normal and insulin-resistant states, independent of insulin [28,29]. In addition, although a peroxisome proliferator response element (PPRE) site in the GLUT4 gene has not been identified [30,31], PPARγ appears to increase GLUT4 [30-33]. In spite of that, the precise underlying mechanism through which sulfonylureas and thiazolidinediones increase glucose uptake in extrapancreatic tissues has not been verified. It is suggested that PPARγ, activated by certain sulfonylureas, induces GLUT4 to be inserted into the plasma membrane through direct interaction of glimepiride with PPARγ molecules or through another indirect mechanism involving a definite signaling pathway that leads to increased glucose uptake.
- Recently, the Food and Drug Administration (FDA) limited the use of rosiglitazone due to increased cardiovascular risk [34]. Sulfonylureas are also known to increase the risk of cardiovascular events [35,36], mostly attributed to their effect on myocardial ATP-sensitive K+ channels. Because some sulfonylureas activate PPARγ, one cannot exclude the possibility that increasing PPARγ activity might also contribute to sulfonylurea's cardiovascular effect.
- Our findings that sulfonylureas induce PPARγ transcriptional activity at clinically relevant concentrations will be helpful for understanding the mechanisms of sulfonylureas in the treatment of subjects with type 2 diabetes mellitus.
DISCUSSION
-
Acknowledgements
- This work was supported by MarineBio21, Ministry of Maritime Affairs and Fisheries, Korea.
ACKNOWLEDGMENTS
- 1. Reaven GM. Banting lecture 1988: role of insulin resistance in human disease. Diabetes 1988;37:1595-1607. ArticlePubMed
- 2. Taylor SI. Deconstructing type 2 diabetes. Cell 1999;97:9-12. ArticlePubMed
- 3. Olefsky JM. Treatment of insulin resistance with peroxisome proliferator-activated receptor gamma agonists. J Clin Invest 2000;106:467-472. ArticlePubMedPMC
- 4. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008;77:289-312. ArticlePubMed
- 5. Panigrahy D, Singer S, Shen LQ, Butterfield CE, Freedman DA, Chen EJ, Moses MA, Kilroy S, Duensing S, Fletcher C, Fletcher JA, Hlatky L, Hahnfeldt P, Folkman J, Kaipainen A. PPARgamma ligands inhibit primary tumor growth and metastasis by inhibiting angiogenesis. J Clin Invest 2002;110:923-932. ArticlePubMedPMC
- 6. Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature 1992;355:446-449. ArticlePubMedPMCPDF
- 7. Renaud JP, Moras D. Structural studies on nuclear receptors. Cell Mol Life Sci 2000;57:1748-1769. ArticlePubMedPDF
- 8. Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 1999;20:649-688. ArticlePubMed
- 9. Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998;93:229-240. ArticlePubMed
- 10. Schopfer FJ, Lin Y, Baker PR, Cui T, Garcia-Barrio M, Zhang J, Chen K, Chen YE, Freeman BA. Nitrolinoleic acid: an endogenous peroxisome proliferator-activated receptor gamma ligand. Proc Natl Acad Sci U S A 2005;102:2340-2345. PubMedPMC
- 11. Lehrke M, Lazar MA. The many faces of PPARgamma. Cell 2005;123:993-999. ArticlePubMed
- 12. Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA. Peroxisome proliferator-activated receptors alpha and gamma are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem 1997;272:3406-3410. PubMed
- 13. Benson SC, Pershadsingh HA, Ho CI, Chittiboyina A, Desai P, Pravenec M, Qi N, Wang J, Avery MA, Kurtz TW. Identification of telmisartan as a unique angiotensin II receptor antagonist with selective PPARgamma-modulating activity. Hypertension 2004;43:993-1002. ArticlePubMed
- 14. Schupp M, Janke J, Clasen R, Unger T, Kintscher U. Angiotensin type 1 receptor blockers induce peroxisome proliferator-activated receptor-gamma activity. Circulation 2004;109:2054-2057. ArticlePubMed
- 15. Fukuen S, Iwaki M, Yasui A, Makishima M, Matsuda M, Shimomura I. Sulfonylurea agents exhibit peroxisome proliferator-activated receptor gamma agonistic activity. J Biol Chem 2005;280:23653-23659. PubMed
- 16. Inukai K, Watanabe M, Nakashima Y, Takata N, Isoyama A, Sawa T, Kurihara S, Awata T, Katayama S. Glimepiride enhances intrinsic peroxisome proliferator-activated receptor-gamma activity in 3T3-L1 adipocytes. Biochem Biophys Res Commun 2005;328:484-490. PubMed
- 17. Arrault A, Rocchi S, Picard F, Maurois P, Pirotte B, Vamecq J. A short series of antidiabetic sulfonylureas exhibit multiple ligand PPARgamma-binding patterns. Biomed Pharmacother 2009;63:56-62. PubMed
- 18. Scarsi M, Podvinec M, Roth A, Hug H, Kersten S, Albrecht H, Schwede T, Meyer UA, Rucker C. Sulfonylureas and glinides exhibit peroxisome proliferator-activated receptor gamma activity: a combined virtual screening and biological assay approach. Mol Pharmacol 2007;71:398-406. ArticlePubMed
- 19. Langtry HD, Balfour JA. Glimepiride: a review of its use in the management of type 2 diabetes mellitus. Drugs 1998;55:563-584. ArticlePubMed
- 20. Malerczyk V, Badian M, Korn A, Lehr KH, Waldhausl W. Dose linearity assessment of glimepiride (Amaryl) tablets in healthy volunteers. Drug Metabol Drug Interact 1994;11:341-357. ArticlePubMed
- 21. von Nicolai H, Brickl R, Eschey H, Greischel A, Heinzel G, Konig E, Limmer J, Rupprecht E. Duration of action and pharmacokinetics of the oral antidiabetic drug gliquidone in patients with non-insulin-dependent (type 2) diabetes mellitus. Arzneimittelforschung 1997;47:247-252. PubMed
- 22. Inukai K, Watanabe M, Nakashima Y, Sawa T, Takata N, Tanaka M, Kashiwabara H, Yokota K, Suzuki M, Kurihara S, Awata T, Katayama S. Efficacy of glimepiride in Japanese type 2 diabetic subjects. Diabetes Res Clin Pract 2005;68:250-257. ArticlePubMed
- 23. Jaber LA, Ducharme MP, Edwards DJ, Slaughter RL, Grunberger G. The influence of multiple dosing and age on the pharmacokinetics and pharmacodynamics of glipizide in patients with type II diabetes mellitus. Pharmacotherapy 1996;16:760-768. ArticlePubMed
- 24. Hiromori Y, Nishikawa J, Yoshida I, Nagase H, Nakanishi T. Structure-dependent activation of peroxisome proliferator-activated receptor (PPAR) gamma by organotin compounds. Chem Biol Interact 2009;180:238-244. PubMed
- 25. Mori RC, Hirabara SM, Hirata AE, Okamoto MM, Machado UF. Glimepiride as insulin sensitizer: increased liver and muscle responses to insulin. Diabetes Obes Metab 2008;10:596-600. ArticlePubMed
- 26. Rosskamp R, Wernicke-Panten K, Draeger E. Clinical profile of the novel sulphonylurea glimepiride. Diabetes Res Clin Pract 1996;31(Suppl):S33-S42. ArticlePubMed
- 27. Haupt A, Kausch C, Dahl D, Bachmann O, Stumvoll M, Haring HU, Matthaei S. Effect of glimepiride on insulin-stimulated glycogen synthesis in cultured human skeletal muscle cells: a comparison to glibenclamide. Diabetes Care 2002;25:2129-2132. PubMed
- 28. Muller G, Wied S, Wetekam EM, Crecelius A, Unkelbach A, Punter J. Stimulation of glucose utilization in 3T3 adipocytes and rat diaphragm in vitro by the sulphonylureas, glimepiride and glibenclamide, is correlated with modulations of the cAMP regulatory cascade. Biochem Pharmacol 1994;48:985-996. ArticlePubMed
- 29. Muller G. The molecular mechanism of the insulin-mimetic/sensitizing activity of the antidiabetic sulfonylurea drug Amaryl. Mol Med 2000;6:907-933. ArticlePubMedPMCPDF
- 30. Fernyhough ME, Okine E, Hausman G, Vierck JL, Dodson MV. PPARgamma and GLUT-4 expression as developmental regulators/markers for preadipocyte differentiation into an adipocyte. Domest Anim Endocrinol 2007;33:367-378. PubMed
- 31. Ezaki O. Regulatory elements in the insulin-responsive glucose transporter (GLUT4) gene. Biochem Biophys Res Commun 1997;241:1-6. ArticlePubMed
- 32. McGowan KM, Long SD, Pekala PH. Glucose transporter gene expression: regulation of transcription and mRNA stability. Pharmacol Ther 1995;66:465-505. ArticlePubMed
- 33. Hamm JK, el Jack AK, Pilch PF, Farmer SR. Role of PPAR gamma in regulating adipocyte differentiation and insulin-responsive glucose uptake. Ann N Y Acad Sci 1999;892:134-145. PubMed
- 34. Kahn BB, McGraw TE. Rosiglitazone, PPARγ, and type 2 diabetes. N Engl J Med 2010;363:2667-2669. ArticlePubMedPMC
- 35. Tzoulaki I, Molokhia M, Curcin V, Little MP, Millett CJ, Ng A, Hughes RI, Khunti K, Wilkins MR, Majeed A, Elliott P. Risk of cardiovascular disease and all cause mortality among patients with type 2 diabetes prescribed oral antidiabetes drugs: retrospective cohort study using UK general practice research database. BMJ 2009;339:b4731ArticlePubMedPMC
- 36. Evans JM, Ogston SA, Emslie-Smith A, Morris AD. Risk of mortality and adverse cardiovascular outcomes in type 2 diabetes: a comparison of patients treated with sulfonylureas and metformin. Diabetologia 2006;49:930-936. ArticlePubMedPDF
REFERENCES
Fig. 1Peroxisome proliferator-activated receptor γ (PPARγ) transcriptional activity by thiazolidinediones (rosiglitazone) and sulfonylureas (glimepiride, gliquidone, and glipizide). Cos 7 cells were transfected with Gal4 tk-Luc, pCMV-β-gal, and pM-PPARγ and treated with rosiglitazone (1 to 10 µM), glimepiride (1 to 300 µM), gliquidone (1 to 100 µM) or glipizide (1 to 500 µM) for 24 hours. β-galactosidase activity was used for normalization of luciferase activity. The luciferase activity of the cells treated with DMSO was set to 1, and the others were expressed as relative values. Data represent the mean±standard error of the mean (SEM) (n=3).
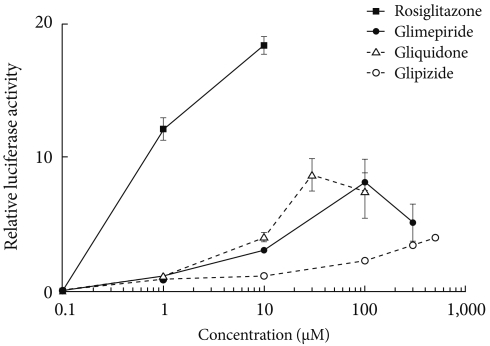

Fig. 2Target regions of sulfonylureas in peroxisome proliferator-activated receptor γ (PPARγ). (A) Schematic diagram of PPARγ constructs. Three constructs from wild type PPARγ (pM-WT) were prepared. pM-ΔLBD lacked the ligand binding domain, pM-AF1 carried only the AF1 region and pM-LBD had only the ligand binding domain. (B) Transcriptional activity according to PPARγ construct. pM-WT, pM-AF1, pM-ΔLBD, or pM-LBD were cotransfected with Gal4 tk-Luc and pCMV-β-gal into COS7 cells. Cells were treated with glimepiride, gliquidone, glipizide or rosiglitazone at indicated doses for 24 hours. The luciferase activity of the cells treated with DMSO after overexpression of PPARγ wild type or its deletions, respectively, was set to 1, and other activities were expressed as relative values. Data represent the mean±standard error of the mean (SEM) (n=5). aP<0.05.
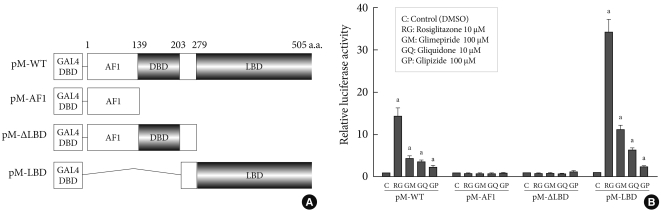

Fig. 3Combination treatments of thiazolidinediones and sulfonylurea. (A) Combination treatments of thiazolidinediones and sulfonylurea. To examine the effects of rosiglitazone in combination with each sulfonylurea on the peroxisome proliferator-activated receptor γ (PPARγ) transcriptional activity, COS7 cells were transfected with Gal4 tk-Luc, pCMV-β-gal, and pM or pM-PPARγ and treated with glimepiride (100 µM), gliquidone (10 µM), or glipizide (100 µM) plus rosiglitazone (1 µM). Data represent the mean±standard error of the mean (SEM) (n=4). aP<0.05. (B) Combination treatment according to sulfonylurea dose. Cos 7 cells were transfected with Gal4 tk-Luc, pCMV-β-gal, or pM-PPARγ and treated with rosiglitazone, glimepiride or gliquidone at the indicated doses for 24 hours. Data represent mean±standard error of the mean (SEM) (n=4). aP<0.05.
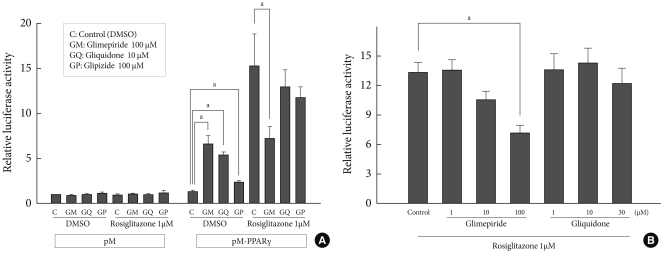

Fig. 4Effects of glimepiride on rosiglitazone-induced glucose uptake. On day 8 of differentiation, 3T3-L1 adipocytes were treated with rosiglitazone (10 µM), glimepiride (100 µM) or both for 48 hours and with insulin for 30 minutes. Glucose uptake was measured using [3H]-deoxyglucose scintillation counting. The glucose uptake value of untreated cells was set to 100, and the others were relative values. Data represent the mean±standard error of the mean (SEM) (n=3). aP<0.05, bP<0.05 compared to the control.
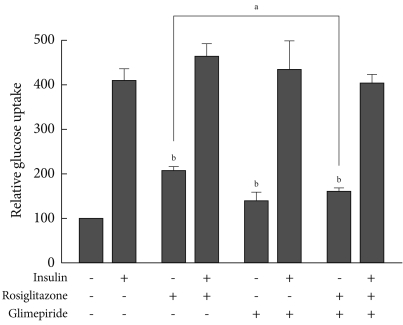

Figure & Data
References
Citations
Citations to this article as recorded by 

- Chitosan-Encapsulated Nano-selenium Targeting TCF7L2, PPARγ, and CAPN10 Genes in Diabetic Rats
Omayma A. R. Abozaid, Sawsan M. El-Sonbaty, Neama M. A. Hamam, Moustafa A. Farrag, Ahmad S. Kodous
Biological Trace Element Research.2023; 201(1): 306. CrossRef - Insights from insulin resistance pathways: Therapeutic approaches against Alzheimer associated diabetes mellitus
Ayesha Fauzi, Ewen Se Thoe, Tang Yin Quan, Adeline Chia Yoke Yin
Journal of Diabetes and its Complications.2023; 37(11): 108629. CrossRef - Novel Sulfonanilide Inhibitors of SHIP2 Enhance Glucose Uptake into Cultured Myotubes
Mika E. A. Berg, Jette-Britt Naams, Laura C. Hautala, Tuomas A. Tolvanen, Jari P. Ahonen, Sanna Lehtonen, Kristiina Wähälä
ACS Omega.2020; 5(3): 1430. CrossRef - Diabetic Theory in Anti-Alzheimer’s Drug Research and Development - Part 1: Therapeutic Potential of Antidiabetic Agents
Agnieszka Jankowska, Anna Wesołowska, Maciej Pawłowski, Grażyna Chłoń-Rzepa
Current Medicinal Chemistry.2020; 27(39): 6658. CrossRef - Moringa concanensis Nimmo extracts ameliorates hyperglycemia-mediated oxidative stress and upregulates PPARγ and GLUT4 gene expression in liver and pancreas of streptozotocin-nicotinamide induced diabetic rats
Brindha Banu Balakrishnan, Kalaivani Krishnasamy, Vijayakumar Mayakrishnan, Arokiyaraj Selvaraj
Biomedicine & Pharmacotherapy.2019; 112: 108688. CrossRef - PPARγ Agonistic Activity of Sulphonylureas
Debjani Banerjee, Harnovdeep Singh Bharaj, Moulinath Banerjee
Endocrine, Metabolic & Immune Disorders - Drug Targets.2019; 19(4): 467. CrossRef - Glimepiride treatment in a patient with type A insulin resistance syndrome due to a novel heterozygous missense mutation in the insulin receptor gene
Zhimin Huang, Juan Liu, Kaka Ng, Xuesi Wan, Lijuan Xu, Xiaoying He, Zhihong Liao, Yanbing Li
Journal of Diabetes Investigation.2018; 9(5): 1075. CrossRef - Arsenic, Cadmium, and Lead Like Troglitazone Trigger PPARγ-Dependent Poly (ADP-Ribose) Polymerase Expression and Subsequent Apoptosis in Rat Brain Astrocytes
Rajesh Kushwaha, Juhi Mishra, Sachin Tripathi, Puneet Khare, Sanghamitra Bandyopadhyay
Molecular Neurobiology.2018; 55(3): 2125. CrossRef - Docosahexaenoic acid up‐regulates both PI3K/AKT‐dependent FABP7–PPARγ interaction and MKP3 that enhance GFAP in developing rat brain astrocytes
Sachin Tripathi, Rajesh Kushwaha, Juhi Mishra, Manoj Kumar Gupta, Harish Kumar, Somali Sanyal, Dhirendra Singh, Sabyasachi Sanyal, Amogh Anant Sahasrabuddhe, Mohan Kamthan, Mohana Krishna Reddy Mudiam, Sanghamitra Bandyopadhyay
Journal of Neurochemistry.2017; 140(1): 96. CrossRef - Antiglycation and cell protective actions of metformin and glipizide in erythrocytes and monocytes
Krishna Adeshara, Rashmi Tupe
Molecular Biology Reports.2016; 43(3): 195. CrossRef - The therapeutic journey of pyridazinone
Wasim Akhtar, M. Shaquiquzzaman, Mymoona Akhter, Garima Verma, Mohemmed Faraz Khan, M. Mumtaz Alam
European Journal of Medicinal Chemistry.2016; 123: 256. CrossRef - Antidiabetic effect of novel benzenesulfonylureas as PPAR-γ agonists and their anticancer effect
Chetna Kharbanda, Mohammad Sarwar Alam, Hinna Hamid, Kalim Javed, Abhijeet Dhulap, Sameena Bano, Yakub Ali
Bioorganic & Medicinal Chemistry Letters.2015; 25(20): 4601. CrossRef - Method Development and Validation of Amlodipine, Gliquidone and Pioglitazone: Application in the Analysis of Human Serum
Agha Zeeshan Mirza, M. Saeed Arayne, Najma Sultana
Analytical Chemistry Letters.2014; 4(5-6): 302. CrossRef - Gliquidone decreases urinary protein by promoting tubular reabsorption in diabetic Goto-Kakizaki rats
Jian-Ting Ke, Mi Li, Shi-Qing Xu, Wen-Jian Zhang, Yong-Wei Jiang, Lan-yun Cheng, Li Chen, Jin-Ning Lou, Wei Wu
Journal of Endocrinology.2014; 220(2): 129. CrossRef - Extension of Drosophila lifespan by cinnamon through a sex-specific dependence on the insulin receptor substrate chico
Samuel E. Schriner, Steven Kuramada, Terry E. Lopez, Stephanie Truong, Andrew Pham, Mahtab Jafari
Experimental Gerontology.2014; 60: 220. CrossRef - Crif1 Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Min Jeong Ryu, Soung Jung Kim, Yong Kyung Kim, Min Jeong Choi, Surendar Tadi, Min Hee Lee, Seong Eun Lee, Hyo Kyun Chung, Saet Byel Jung, Hyun-Jin Kim, Young Suk Jo, Koon Soon Kim, Sang-Hee Lee, Jin Man Kim, Gi Ryang Kweon, Ki Cheol Park, Jung Uee Lee, Yo
PLoS Genetics.2013; 9(3): e1003356. CrossRef - Glimepiride attenuates Aβ production via suppressing BACE1 activity in cortical neurons
Feiyang Liu, Yijin Wang, Ming Yan, Luyong Zhang, Tao Pang, Hong Liao
Neuroscience Letters.2013; 557: 90. CrossRef - Pharmacologic agents for type 2 diabetes therapy and regulation of adipogenesis
A. Cignarelli, F. Giorgino, R. Vettor
Archives of Physiology and Biochemistry.2013; 119(4): 139. CrossRef - Labisia pumilaUpregulates Peroxisome Proliferator-Activated Receptor Gamma Expression in Rat Adipose Tissues and 3T3-L1 Adipocytes
Fazliana Mansor, Harvest F. Gu, Claes-Göran Östenson, Louise Mannerås-Holm, Elisabet Stener-Victorin, Wan Nazaimoon Wan Mohamud
Advances in Pharmacological Sciences.2013; 2013: 1. CrossRef - Protocol for effective differentiation of 3T3-L1 cells to adipocytes
Katja Zebisch, Valerie Voigt, Martin Wabitsch, Matthias Brandsch
Analytical Biochemistry.2012; 425(1): 88. CrossRef

 KDA
KDA PubReader
PubReader Cite
Cite