Lack of Evidence of the Role of APOA5 3’UTR Polymorphisms in Iranian Children and Adolescents with Metabolic Syndrome
Article information
Abstract
Background
Metabolic syndrome (MetS) is a complex and multifactorial disorder characterized by insulin resistance, dyslipidaemia, hyperglycemia, abdominal obesity, and elevated blood pressure. The apolipoprotein A5 (APOA5) gene variants have been reported to correlate with two major components of MetS, including low levels of high density lipoprotein cholesterol (HDL-C) and high levels of triglyceride. In the present study, we explored the associations between five single nucleotide polymorphisms (SNPs) of APOA5 gene and the MetS risk.
Methods
In a case-control design, 120 Iranian children and adolescents with/without MetS were genotyped by polymerase chain reaction-sequencing for these SNPs. Then, we investigated the association of SNPs, individually or in haplotype constructs, with MetS risk.
Results
The rs34089864 variant and H1 haplotype (harboring the two major alleles of rs619054 and rs34089864) were associated with HDL-C levels. However, there was no significant association between different haplotypes/individual SNPs and MetS risk.
Conclusion
These results presented no association of APOA5 3’UTR SNPs with MetS. Further studies, including other polymorphisms, are required to investigate the involvement of APOA5 gene in the genetic susceptibility to MetS in the pediatric age group.
INTRODUCTION
Metabolic syndrome (MetS) is clustering of several cardiovascular disease risk factors such as dyslipidaemia, hyperglycemia, abdominal obesity and elevated blood pressure [12]. Several complex etiological mechanisms such as genetics and environmental factors are considered to be related to MetS [2].
MetS is considerably influenced by genetic factors. Apolipoprotein A5 (APOA5) gene is located on chromosome 11q23 in the APOA1-C3-A4 gene cluster encoding the APOAV protein with 343 amino acids. APOA5 plays an important role in lipid metabolism and homeostasis. Moreover, plasma APOA5 levels have been considered to reduce plasma triglyceride (TG) and increase high density lipoprotein cholesterol (HDL-C) concentrations. Several single nucleotide polymorphisms (SNPs) in the APOA5 gene have been shown to be associated with increased TG levels [345].
MicroRNAs (miRNAs) are a group of non-protein-coding small RNAs regulating gene expression post-transcriptionally [67]. SNPs in miRNA target site (miR-TS-SNP) may affect or impair binding of miRNAs [8] and considerably affect diverse biological functions, including disease susceptibilities (cancers, Parkinson disease, osteoporosis, diabetes, and hypertension etc.) in humans [78910].
In the current study, a panel of five SNPs (rs619054, rs369740771, rs376976905, rs192708363, and rs34089864) within APOA5 gene was selected to study their association with MetS. Rs619054 variant is significantly associated with HDL, low density lipoprotein cholesterol (LDL-C) and TG levels [111213]. This polymorphism disrupts hsa-mir-28-5p, hsa-mir-708, and hsa-mir-3139 binding sites, and creates a new binding site for hsa-mir-320e. Rs34089864 variant destroys hsa-mir-4511 and hsa-mir-498 binding sites, and creates new binding sites for hsa-mir-4740-5p, hsa-mir-561-5p, and hsa-mir-3169.
The main aim of our case-control study was to evaluate the genotype and allele frequencies of two variants in the three prime untranslated regions (3’UTRs) of APOA5 gene, including rs619054 (c.⋆31C>T or 1764C>T) and rs34089864 (c.⋆76C>T or c.1177C>T) to find any potential associations with MetS and multiple features of this syndrome. Furthermore, we determined the haplotype frequencies and the associations of the five SNPs at the APOA5 locus with the risk of MetS.
METHODS
Study population
This case-control study included 57 patients (24 males and 33 females: mean age, 12.40±0.26 years [range, 9 to 18 years]) and 63 controls (28 males and 35 females: mean age, 13.39±0.38 years [range, 8 to 18 years]) from children and adolescent of Isfahan (a central city in Iran) [14]. According to the modified Adult Treatment Panel III (ATP III) criteria [15], the MetS was defined as clustering of the following three or more factors in one person (1) fasting TG ≥100 mg/dL; (2) HDL-C <50 mg/dL; (3) waist circumference >75th percentile for age and gender in the studied population; (4) systolic blood pressure/diastolic blood pressure >90th percentile for gender, age, height [16]; and (5) fasting blood sugar ≥100 mg/dL. An oral assent was obtained from participants, and all parents signed a written informed consent after explaining the study objectives and details. Blood samples were collected in two separate tubes for DNA analysis and biochemical tests.
DNA genotyping
DNA was extracted from blood leukocytes using the Diatome kit (Isogen Laboratory, Moscow, Russia) according to the manufacturer's recommendations. Genotyping for the 5 miR-TS-SNP at APOA5 gene was carried out by polymerase chain reaction (PCR) and direct sequencing. Primers A5 forward (5′-CTGCCCAGGCCCATTCCCA-3′) and A5 reverse (5′-GAC CTTCCACCCTCCACCCA-3′) were used to amplify the target region of DNA. To amplify the 104 bp fragment of the APOA5 gene, the following conditions were used: a 2.5 minutes initial denaturation at 95℃ was followed by 40 cycles of 50 seconds at 95℃; 70 seconds at 68℃; and 70 seconds at 72℃; the final extension occurred at 72℃ for 15 minutes. The PCR reagent mix was as follows: Taq plus DNA polymerase, 0.25 µL (5 U/µL) (KBC, Terhran, Iran); 10× buffer, 2.5 µL; MgCl2 (50 mM), 0.75 µL; deoxynucleoside triphosphate (40 mM), 0.5 µL; DNA (200 ng/µL), 2 µL; 0.5 µL of the appropriate specific primer pairs (10 pmol/µL) and water in a final volume of 25 mL.
MiR-TS-SNPs selection
We used two different methods to select SNPs in miRNA seed regions. One approach was based on identifying the previously known miR-TS-SNPs. PolymiRTS (http://compbio.uthsc.edu/miRSNP/) [17], MirSNP (http://202.38.126.151/hmdd/mirsnp/search/) and miRNASNP (http://www.bioguo.org/miRNASNP/) are three examples of databases using this approach. In the other approach, we first predicted miRNA target sites within the 3’UTRs of the APOA5 gene by using the miRanda (http://www.microrna.org/microrna/getMirnaForm.do) [18] and TargetScan (http://www.targetscan.org/vert_50/) [19]. Then, a list of all SNPs occurring in the 3’UTRs of the APOA5 gene was obtained from the National Center for Biotechnology Information (NCBI) dbSNP database (http://www.ncbi.nlm.nih.gov/). The overlapping SNPs between these two steps were selected for final genotyping (Table 1).
Statistical analysis
Agreement of genotype frequencies with Hardy-Weinberg equilibrium (HWE) was determined using a modified chi-square test. Allele frequencies were estimated by gene counting, and the differences between case/control statuses were examined by a chi-square test. Owing to a limited number of individuals who were homozygous for the minor alleles of rs619054 and rs34089864, the genotypes for these two polymorphisms were grouped as T-carriers (TC+TT) and non-carriers (CC). One-way analysis of variance was used to compare mean values of quantitative traits across TC+TT/CC groups. The association between APOA5 polymorphisms and MetS was assessed by simple and multivariable adjusted odds ratio (OR) corresponding to a 95% confidence interval according to the logistic regression analysis. Multivariate regression analysis models were adjusted for age and gender. Constructed haplotypes, haplotype tags, and standardized linkage disequilibrium (LD), D’, and correlation, r2, and coefficients for all pairs of polymorphisms were assessed with Haploview software version 4.2 (Broad Institute, Cambridge, MA, USA) [20]. Furthermore, we used a four-gamete rule to define the haplotype blocks. Haplotype frequencies were inferred using the expectation-maximization by using HAPSTAT version 3.0 software (University of North Carolina, Chapel Hill, NC, USA; http://www.bios.unc.edu/*lin/hapstat) [2122]. Using the chi-square test, the potential difference between haplotype frequencies of case and control subjects was determined. Statistical analyses were performed using the SPSS version 16.0 statistical software (SPSS Inc., Chicago, IL, USA). All data were presented in the format of mean±standard error of the mean, and P values lower than 0.05 were considered to be statistically significant.
RESULTS
Table 2 presents genotyping results and allele frequencies of the two analyzed APOA5 variants (rs619054 and rs34089864). No significant difference was observed between the MetS and control groups for both genotype and allele frequencies of rs619054 and rs34089864. Moreover, none of the variants violated the HWE (data not shown).

Genotype distributions and allele frequencies of miR-TS-SNPs in controls and patients with metabolic syndrome
Tables 3 and 4 represent stratification of the biochemical laboratory parameters of the control and MetS subjects according to different genotype groups of the analyzed variants. For rs619054, except for a borderline significant difference observed for insulin levels between genotype groups of control subjects, there was no statistically significant difference between other biochemical factors. For rs34089864, the HDL-C level of the CC group was significantly lower than that in the heterozygotes harbouring cytidine and thymine bases (CT) control group. Moreover, a borderline significant difference in the insulin levels between genotype groups was shown in both case and control individuals.

Biochemical characteristics of subjects with metabolic syndrome and controls separated according to APOA5 rs619054 different genotype groups

Biochemical characteristics of subjects with metabolic syndrome and controls separated according to APOA5 rs34089864 different genotype groups
The ORs were evaluated for allelic and genotypic models (Table 5). Variant rs619054 demonstrated inconsequential elevated risks for MetS in both models, but both ORs were not statistically significant. Having a C allele or CC genotype in the position of rs34089864 conferred an increased risk for MetS; however, again the risk was not statistically significant.

Logistic regression models for the association between APOA5 rs619054, rs34089864 and metabolic syndrome risk
Haplotypes were inferred using all five studied variants of the APOA5 gene (rs619054, rs369740771, rs376976905, rs1927 08363, and rs34089864). Rs619054 and rs34089864 were in LD with each other (D’=1, r2=0.02, LOD [logarithm of the odds to the base 10]=0.37). Three haplotypes were identified: H1, H2, and H3. These haplotypes are tagged by the genotypes of rs619054 and rs34089864 variants. Fig. 1 presents haplotype frequencies. Further analyses showed no significant difference between haplotype frequencies according to the disease status.
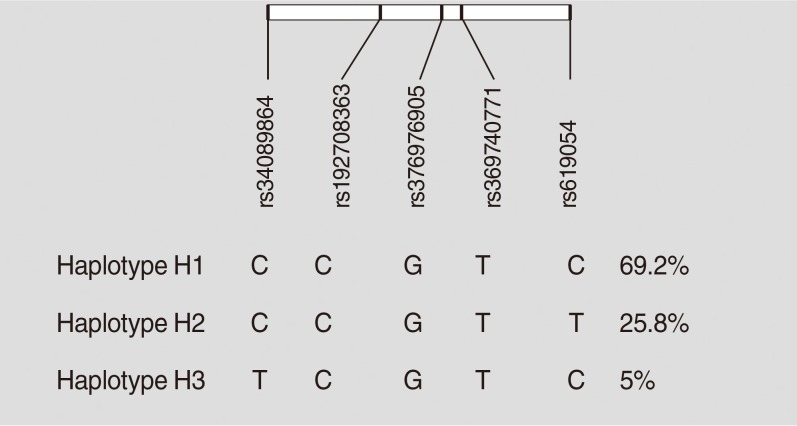
Haplotype characterization and frequencies of apolipoprotein A5 (APOA5) polymorphisms in pooled metabolic syndrome and control groups constructed by Haploview software.
Based on stratification for different haplotypes (Table 6), MetS individuals with H3 showed significant higher levels of TG compared to other haplotypes (P=0.04). A significant increase in the HDL-C levels was also observed in controls with H3 compared to the other haplotypes (P=0.01).

APOA5 haplotypes identified in the case and control groups and their correlation with anthropometric and biochemical parameters
Table 7 shows the association between different constructed haplotypes and risk of MetS. Although not statistically significant, individuals with H3 had the most increased risk for MetS compared to those harboring H2 before and after adjustment for age and gender.
DISCUSSION
Elevated TG and lowered HDL-C levels are the major factors among patients with MetS in Iranian children and adolescents [15]. Genetic variants of APOA5 gene have been associated with the levels of TG and HDL-C in various ethnicities [23242526]. In the current study, we have sequenced the 3’UTR of APOA5 gene in groups of 63 patients and 59 healthy controls to find potential associations with ATP III-defined MetS, and its major components in the Iranian children and adolescent population. The results showed that the alleles and genotype frequency of the two common studied SNPs (rs619054 and rs34089864) were not significantly varied in the two groups.
In one case-control study population in China, Kao et al. [27] demonstrated that c.1177C>T (rs34089864) genotype and allele frequencies did not differ between the hypertriglyceridemic and control groups. In agreement with results obtained by Kao et al. [27], we also found no significant associations between serum TG levels and c.1177C>T genotypes.
Our results demonstrated that CC genotype of variant rs34089864 was associated with lower HDL-C levels in MetS individuals. Conversely, the data showed no association between the rs619054 SNP and MetS components. In one case-control population, Pare et al. [12] showed that the rs619054 was not associated with TG, LDL-C, and HDL-C levels. Using joint analysis of the family and case-control populations, they could find a significant association with TG, but neither with LDL-C nor with HDL-C levels. Therefore, in one large population study, Grallert et al. [28] demonstrated that no significant associations were observed between rs619054, HDL-C, and LDL-C, or total cholesterol levels. In contrast, Jorgensen et al. [11] reported that CC genotype of rs619054 SNP was associated with both HDL-C and TG, but not with LDL-C levels. Therefore, the observed discrepancies between the studies may be attributed to the different race/ethnicity and age distributions of the studied populations.
Consistent with the results previously reported by Grallert et al. [28] who showed that rs619054 was not significantly associated with MetS, we found no significant association between any of the two studied major variants and MetS risk.
We also performed a haplotype analysis constructed using five SNPs in the 3’UTR of APOA5 gene. Similar to what found in Copenhagen populations [11], rs619054 and rs34089864 were in LD in our analysis.
Among the three constructed haplotypes, H1, H2, and H3, we found a significant association between H3 haplotype, increased HDL-C and TG levels, but not with MetS risk. In the same vein, H1 haplotype (harboring the two major alleles of rs619054 and rs34089864) was linked to decreased HDL-C levels. Our data are in consistent with those obtained by Pullinger et al. [29] where they constructed haplotypes with 11 APOA5 gene SNPs, including rs619054 and rs34089864 in the carriers of the c.553T polymorphism and reported that a haplotype containing the major alleles of variant rs619054 and rs34089864 was significantly associated with lowered levels of HDL-C.
In conclusion, our study explored the association between haplotypes of five SNPs located at APOA5 3’UTR and MetS entity as well as its major components risk. Our results showed that the constructed haplotypes might not be a major risk factor for the MetS entity in Iranian children and adolescents. However, our results demonstrated that the rs34089864 variant and H1 haplotype (harboring the two major alleles of rs619054 and rs34089864) were associated with HDL-C levels.
ACKNOWLEDGMENTS
This study was supported in part by a research grant from the Pediatric Inherited Diseases Research Center, Research Institute for Primordial Prevention of Non-communicable Disease, Isfahan University of Medical Sciences, Isfahan, Iran. The authors thank Prof. Mahin Hashemipour, Ms. Ghazaleh Fatemi, and Ms. Hajar Miranzadeh for their great help in the sample collection and experiments optimization steps.
Notes
CONFLICTS OF INTEREST: No potential conflict of interest relevant to this article was reported.









